Very present in Archean seawater, nickel is now found at nanomolar concentrations in most environments. Thus, several nickel enzymes have ancestral origin. They have appeared on Earth in environments rich in iron, nickel, H2, CO or CO2 and their presence is essential in various processes. Among them, carbon monoxide dehydrogenase (CODH) reversibly catalyzes the oxidation of CO to CO2. It plays a central role in the global carbon cycle in anaerobic microorganisms. Interest in nickel enzymes such as CODH has grown in recent years due to the search for new energies and the development of new methods of CO2 sequestration. From a fundamental point of view, understanding the biosynthetic pathway of these metalloenzymes has become an ambitious topic in the field of bio-inorganic chemistry.
CODH active site is unique in biology and its biosynthesis intrigues both chemists and biologists. The crystal structure of CODH revealed the nature of its active site, consisting of iron, sulfur and nickel
[1]. The key step in enzyme activation is the insertion of nickel into the enzyme. This step requires the intervention of three accessory proteins, CooC, CooJ and CooT; the last two have only been described in the bacterium
Rhodospirillum rubrum.
Recently, the
Biocatalysis
team of the Chemistry and Biology of Metals laboratory revealed through phylogenetic analysis the existence of 111 CooT homologs related to nickel metabolism in anaerobic bacteria and archaea. The signature of these proteins is the strict conservation of cysteine at position 2. A partially conserved histidine is also present in the C-terminal region of the protein. The biophysical and structural characterization of CooT from
R. rubrum revealed that the protein is able to specifically bind one nickel ion per dimer, and this only thanks to cysteine 2
[2]. Subsequently, the researchers of this team, in collaboration with the
BioMet team from the same laboratory, identified and characterized the CooT protein synthesized by another bacterium (Carboxydothermus hydrogenoformans), revealing a second nickel-bindng mode that involves both the cysteine and the histidine
[3]. The researchers have identified a novel family of nickel proteins whose members have two distinct nickel coordination motifs.
This study deciphers the biosynthesis of CODH to understand how the enzyme is matured and its active site formed.
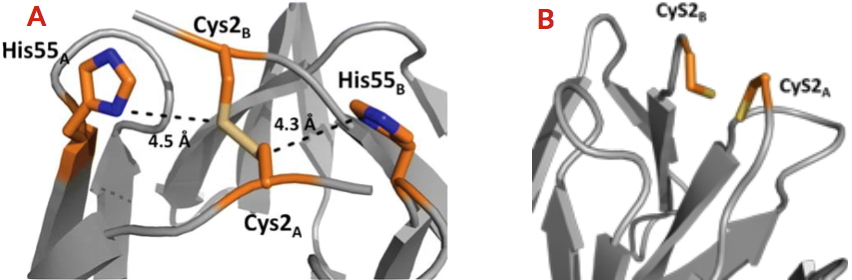
A - Nickel-binding site of CooT from Carboxydothermus hydrogenoformans.
B - Nickel-binding site of CooT from Rhodospirillum rubrum.